DNA microarray technology allows for the simultaneous analysis of thousands of genes by hybridizing cDNA to known probes, offering a cost-effective method for gene expression profiling. RNA-Seq provides a more comprehensive and precise measurement of transcriptomes by sequencing cDNA fragments, enabling the detection of novel transcripts and alternative splicing events. In biotechnology pet applications, RNA-Seq outperforms DNA microarrays by delivering higher sensitivity and greater dynamic range for studying gene expression changes linked to pet health and genetics.
Table of Comparison
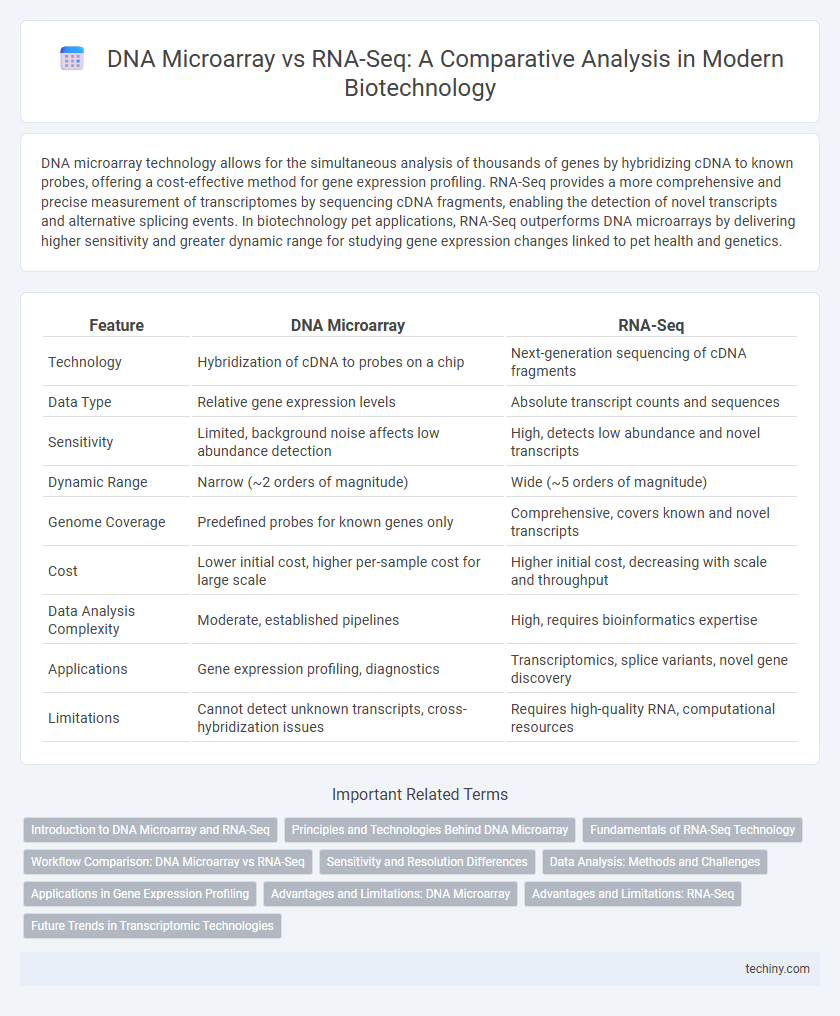
| Feature | DNA Microarray | RNA-Seq |
|---|---|---|
| Technology | Hybridization of cDNA to probes on a chip | Next-generation sequencing of cDNA fragments |
| Data Type | Relative gene expression levels | Absolute transcript counts and sequences |
| Sensitivity | Limited, background noise affects low abundance detection | High, detects low abundance and novel transcripts |
| Dynamic Range | Narrow (~2 orders of magnitude) | Wide (~5 orders of magnitude) |
| Genome Coverage | Predefined probes for known genes only | Comprehensive, covers known and novel transcripts |
| Cost | Lower initial cost, higher per-sample cost for large scale | Higher initial cost, decreasing with scale and throughput |
| Data Analysis Complexity | Moderate, established pipelines | High, requires bioinformatics expertise |
| Applications | Gene expression profiling, diagnostics | Transcriptomics, splice variants, novel gene discovery |
| Limitations | Cannot detect unknown transcripts, cross-hybridization issues | Requires high-quality RNA, computational resources |
Introduction to DNA Microarray and RNA-Seq
DNA microarray technology enables the simultaneous analysis of thousands of gene expressions by hybridizing labeled cDNA to probes fixed on a solid surface, allowing for high-throughput gene expression profiling. RNA-Seq, or RNA sequencing, uses next-generation sequencing to capture the complete transcriptome, providing detailed insights into gene expression levels, splice variants, and novel transcripts with higher sensitivity and dynamic range compared to DNA microarrays. Both techniques are crucial in functional genomics but differ in sensitivity, quantification accuracy, and the ability to detect unknown transcripts.
Principles and Technologies Behind DNA Microarray
DNA microarray technology utilizes thousands of immobilized DNA probes on a solid surface to hybridize with fluorescently labeled cDNA or RNA samples, enabling simultaneous measurement of gene expression levels. This platform relies on complementary base pairing and fluorescence detection, providing a high-throughput approach to analyze gene activity across the genome. The hybridization intensity correlates with transcript abundance, making DNA microarray a powerful tool for comparative transcriptomics and gene expression profiling.
Fundamentals of RNA-Seq Technology
RNA-Seq technology involves sequencing cDNA synthesized from RNA molecules, enabling comprehensive analysis of transcriptomes with high sensitivity and dynamic range. Unlike DNA microarrays that rely on hybridization to predefined probes, RNA-Seq detects both known and novel transcripts, quantifying gene expression levels directly through read counts. This sequencing-based approach allows for precise identification of alternative splicing events, allele-specific expression, and transcript isoforms, providing deeper insight into cellular functions and disease mechanisms.
Workflow Comparison: DNA Microarray vs RNA-Seq
DNA microarray workflow involves hybridizing fluorescently labeled cDNA to predefined probes on a chip, enabling simultaneous analysis of thousands of known gene sequences with relative expression levels. RNA-Seq workflow includes converting RNA into cDNA, fragmenting it, and performing high-throughput sequencing, offering comprehensive transcriptome profiling with detection of novel transcripts and splice variants. RNA-Seq provides higher sensitivity, dynamic range, and data depth compared to DNA microarrays, while DNA microarrays offer a more cost-effective and faster approach for targeted gene expression studies.
Sensitivity and Resolution Differences
DNA microarray technology detects known gene sequences with moderate sensitivity, while RNA-Seq provides higher sensitivity by identifying both known and novel transcripts. RNA-Seq offers superior resolution, enabling detection of low-abundance transcripts and single-nucleotide variations that microarrays cannot accurately capture. The comprehensive coverage and quantitative accuracy of RNA-Seq make it the preferred method for detailed gene expression analysis in biotechnology research.
Data Analysis: Methods and Challenges
DNA microarray data analysis relies on fluorescence intensity measurements to quantify gene expression, requiring rigorous normalization techniques such as RMA or MAS5 to correct for technical variations. RNA-Seq offers greater sensitivity and dynamic range, utilizing sequence alignment and transcript quantification tools like STAR and HTSeq, but challenges include handling large datasets, mapping biases, and differential expression statistical modeling with DESeq2 or edgeR. Both methods demand sophisticated bioinformatics pipelines and statistical expertise to accurately interpret gene expression profiles and biological significance.
Applications in Gene Expression Profiling
DNA microarray and RNA-Seq are both powerful technologies for gene expression profiling, with RNA-Seq offering higher sensitivity and the ability to detect novel transcripts and alternative splicing events. RNA-Seq provides quantitative data across a broader dynamic range, enabling more accurate measurement of low-abundance transcripts compared to DNA microarrays. This advantage makes RNA-Seq particularly valuable for comprehensive transcriptomic studies in disease research, biomarker discovery, and functional genomics.
Advantages and Limitations: DNA Microarray
DNA microarray technology enables simultaneous analysis of thousands of gene expressions by hybridizing cDNA to pre-designed probes, offering cost-effective and high-throughput profiling. However, it is limited by dependency on prior knowledge of gene sequences, lower sensitivity for low-abundance transcripts, and reduced dynamic range compared to RNA-Seq. Cross-hybridization and background noise can also affect data accuracy, making it less suitable for novel transcript detection or comprehensive transcriptome analysis.
Advantages and Limitations: RNA-Seq
RNA-Seq offers superior sensitivity and dynamic range compared to DNA microarrays, enabling the detection of novel transcripts, splice variants, and low-abundance genes without prior knowledge of the genome. The technique provides quantitative data with higher resolution, facilitating comprehensive transcriptome profiling and differential expression analysis in diverse biological samples. However, RNA-Seq is limited by higher costs, greater computational demands for data analysis, and potential biases introduced during library preparation and sequencing.
Future Trends in Transcriptomic Technologies
Emerging trends in transcriptomic technologies emphasize the increasing adoption of RNA-Seq over DNA microarray due to RNA-Seq's higher sensitivity, broader dynamic range, and ability to detect novel transcripts and alternative splicing events. Advanced single-cell RNA-Seq platforms and spatial transcriptomics are driving innovations that enable comprehensive gene expression profiling at unprecedented resolution and tissue context. Integration of artificial intelligence and machine learning algorithms enhances data analysis, enabling more accurate interpretation of complex transcriptomic datasets and accelerating discoveries in functional genomics.
DNA microarray vs RNA-Seq Infographic
 techiny.com
techiny.com