In situ Hybridization (ISH) allows for the localization of specific nucleic acid sequences within fixed tissues or cells by using labeled complementary probes, providing spatial information about gene expression. Fluorescence In Situ Hybridization (FISH) enhances this technique by employing fluorescently labeled probes, enabling high-resolution visualization of genetic material under a fluorescence microscope. FISH is particularly valuable for detecting chromosomal abnormalities, gene mapping, and identifying microbial pathogens with greater sensitivity and specificity compared to traditional ISH.
Table of Comparison
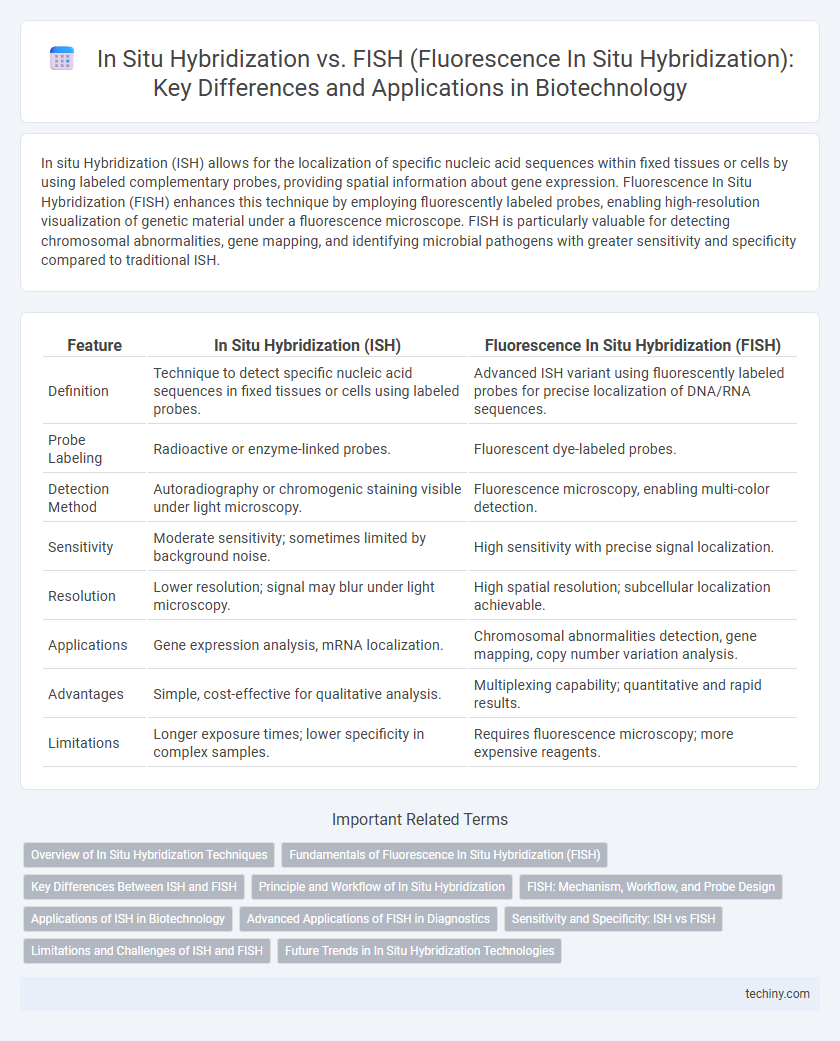
| Feature | In Situ Hybridization (ISH) | Fluorescence In Situ Hybridization (FISH) |
|---|---|---|
| Definition | Technique to detect specific nucleic acid sequences in fixed tissues or cells using labeled probes. | Advanced ISH variant using fluorescently labeled probes for precise localization of DNA/RNA sequences. |
| Probe Labeling | Radioactive or enzyme-linked probes. | Fluorescent dye-labeled probes. |
| Detection Method | Autoradiography or chromogenic staining visible under light microscopy. | Fluorescence microscopy, enabling multi-color detection. |
| Sensitivity | Moderate sensitivity; sometimes limited by background noise. | High sensitivity with precise signal localization. |
| Resolution | Lower resolution; signal may blur under light microscopy. | High spatial resolution; subcellular localization achievable. |
| Applications | Gene expression analysis, mRNA localization. | Chromosomal abnormalities detection, gene mapping, copy number variation analysis. |
| Advantages | Simple, cost-effective for qualitative analysis. | Multiplexing capability; quantitative and rapid results. |
| Limitations | Longer exposure times; lower specificity in complex samples. | Requires fluorescence microscopy; more expensive reagents. |
Overview of In Situ Hybridization Techniques
In situ hybridization (ISH) techniques enable precise localization of specific nucleic acid sequences within fixed tissues or cells, utilizing labeled complementary DNA or RNA probes. Fluorescence in situ hybridization (FISH) is a variant of ISH that employs fluorescent probes to detect and visualize genetic material with high sensitivity and spatial resolution. Both methods are pivotal for gene mapping, transcript localization, and chromosomal analysis in biotechnology research.
Fundamentals of Fluorescence In Situ Hybridization (FISH)
Fluorescence In Situ Hybridization (FISH) employs fluorescent probes that bind to specific DNA sequences, enabling precise localization of genetic material within chromosomes or cells. This technique enhances spatial resolution compared to traditional In Situ Hybridization by using fluorophores for direct visualization under a fluorescence microscope. FISH is widely utilized in genetic mapping, diagnosis of chromosomal abnormalities, and detection of gene expression at the cellular level.
Key Differences Between ISH and FISH
In situ hybridization (ISH) uses labeled complementary DNA or RNA probes to detect specific nucleic acid sequences within fixed tissues or cells, providing spatial gene expression data, while fluorescence in situ hybridization (FISH) employs fluorescently labeled probes for enhanced sensitivity and visualization of genetic material under fluorescence microscopy. FISH allows for multiplexing through distinct fluorophores, enabling simultaneous detection of multiple targets and chromosomal aberrations, which is less feasible in traditional ISH. The main differences lie in probe labeling, signal detection methods, and the quantitative precision of gene mapping achievable with FISH compared to ISH.
Principle and Workflow of In Situ Hybridization
In situ hybridization (ISH) is a technique used to detect specific nucleic acid sequences within fixed tissues or cells by using labeled complementary RNA or DNA probes that hybridize to target sequences. The workflow involves sample fixation, permeabilization, probe hybridization under controlled conditions, and subsequent detection via chromogenic or fluorescent methods to visualize the hybridized probes. Unlike FISH, which specifically employs fluorescently labeled probes for direct visualization, ISH can utilize a broader range of labels, allowing for versatile applications in gene expression analysis and localization.
FISH: Mechanism, Workflow, and Probe Design
Fluorescence In Situ Hybridization (FISH) employs fluorescently labeled DNA probes to bind complementary sequences within intact cells, enabling the detection and localization of specific genetic material at the chromosomal or cellular level. The FISH workflow comprises sample fixation, denaturation, probe hybridization, washing to remove unbound probes, and fluorescence microscopy analysis to visualize hybridization signals. Probe design for FISH demands high specificity and stability, often involving synthetic oligonucleotides labeled with fluorophores tailored to target unique DNA or RNA sequences, optimizing binding affinity and reducing background fluorescence.
Applications of ISH in Biotechnology
In situ hybridization (ISH) enables precise localization of specific nucleic acid sequences within intact tissues, facilitating the study of gene expression patterns crucial for developmental biology and pathology. ISH is extensively applied in cancer research to detect oncogene activation and chromosomal abnormalities at the cellular level, aiding in diagnosis and targeted therapy development. Compared to fluorescence in situ hybridization (FISH), ISH is valuable for visualizing RNA transcripts in fixed samples, making it essential for functional genomics and transcriptomics in biotechnology.
Advanced Applications of FISH in Diagnostics
Fluorescence In Situ Hybridization (FISH) offers enhanced sensitivity and specificity over conventional in situ hybridization by using fluorescent probes to detect chromosomal abnormalities and gene mutations in clinical diagnostics. Advanced applications of FISH include identifying cancer-associated gene rearrangements, monitoring minimal residual disease, and detecting microbial pathogens in complex samples, providing rapid and accurate diagnostic results. High-resolution imaging and multiplexing capabilities enable simultaneous visualization of multiple genetic targets, making FISH indispensable in personalized medicine and cytogenetic analysis.
Sensitivity and Specificity: ISH vs FISH
In situ Hybridization (ISH) provides broad detection of nucleic acids with moderate sensitivity and specificity, making it suitable for general gene expression analysis. Fluorescence In Situ Hybridization (FISH) enhances sensitivity and specificity by using fluorescent probes, allowing precise localization and quantification of specific DNA or RNA sequences at the cellular level. FISH's higher resolution and multiplexing capability surpass ISH in detecting low-abundance targets and distinguishing closely related genetic variants.
Limitations and Challenges of ISH and FISH
In situ Hybridization (ISH) faces limitations such as lower sensitivity and specificity compared to FISH, leading to potential background noise and false-positive signals in detecting nucleic acid sequences. FISH overcomes some sensitivity issues but is challenged by its complexity, high cost, and the requirement for fluorescent microscopy, which restricts its accessibility in routine diagnostics. Both techniques require careful probe design and optimization to avoid cross-hybridization and ensure accurate localization of target sequences within cells or tissues.
Future Trends in In Situ Hybridization Technologies
Emerging trends in in situ hybridization technologies emphasize increased multiplexing capabilities and enhanced spatial resolution, driven by advances in probe design and imaging systems. Integration with artificial intelligence and machine learning algorithms enables more accurate analysis and interpretation of complex genetic and epigenetic data within cellular contexts. Development of non-toxic, highly sensitive fluorescent probes and streamlined protocols aims to expand applications in clinical diagnostics and personalized medicine.
In situ Hybridization vs FISH (Fluorescence In Situ Hybridization) Infographic
 techiny.com
techiny.com