Metagenomics analyzes genetic material directly from environmental samples, providing comprehensive insights into microbial communities without the need for cultivation. Single-cell genomics isolates and sequences the genome of individual cells, enabling detailed study of specific microorganisms and their unique functions. Both approaches enhance our understanding of microbial diversity and interaction, crucial for advancing biotechnology applications in pet health and diagnostics.
Table of Comparison
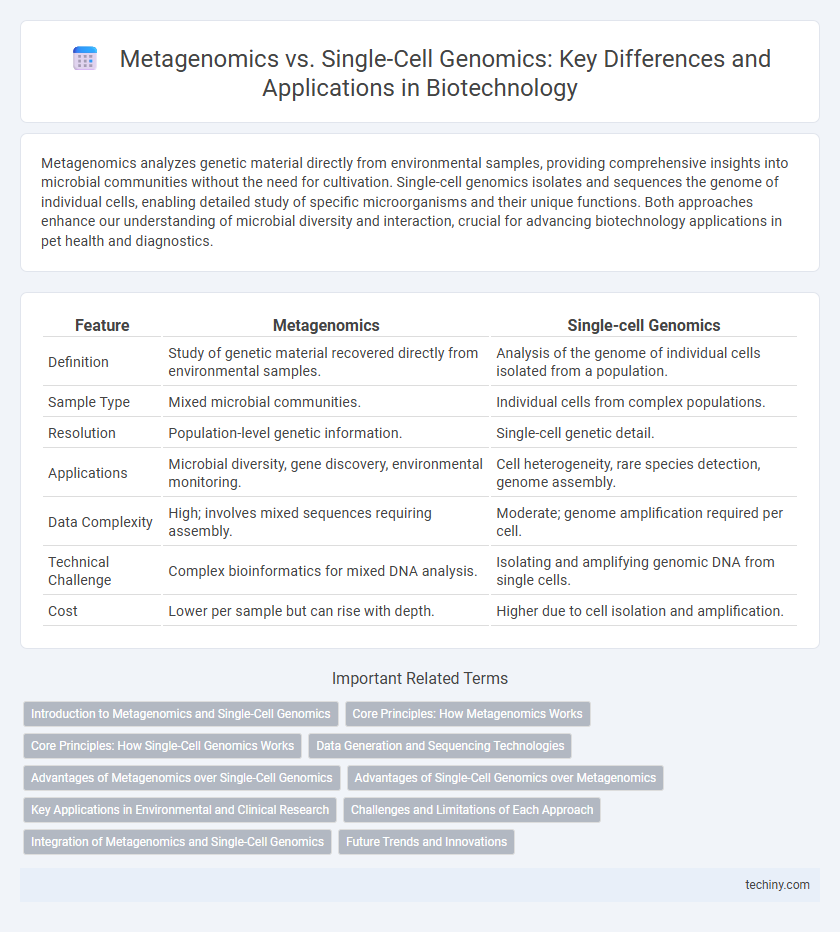
| Feature | Metagenomics | Single-cell Genomics |
|---|---|---|
| Definition | Study of genetic material recovered directly from environmental samples. | Analysis of the genome of individual cells isolated from a population. |
| Sample Type | Mixed microbial communities. | Individual cells from complex populations. |
| Resolution | Population-level genetic information. | Single-cell genetic detail. |
| Applications | Microbial diversity, gene discovery, environmental monitoring. | Cell heterogeneity, rare species detection, genome assembly. |
| Data Complexity | High; involves mixed sequences requiring assembly. | Moderate; genome amplification required per cell. |
| Technical Challenge | Complex bioinformatics for mixed DNA analysis. | Isolating and amplifying genomic DNA from single cells. |
| Cost | Lower per sample but can rise with depth. | Higher due to cell isolation and amplification. |
Introduction to Metagenomics and Single-Cell Genomics
Metagenomics involves the comprehensive analysis of genetic material recovered directly from environmental samples, enabling the study of microbial communities without the need for culturing individual organisms. Single-cell genomics, by contrast, isolates and sequences the genome of individual cells, providing detailed insights into cellular heterogeneity and rare taxa within complex populations. Both approaches enhance understanding of microbial diversity, with metagenomics offering community-wide functional profiles and single-cell genomics revealing organism-specific genomic information.
Core Principles: How Metagenomics Works
Metagenomics involves sequencing genetic material directly from environmental samples, allowing analysis of complex microbial communities without the need for culturing individual organisms. This approach uses high-throughput DNA sequencing combined with bioinformatics tools to assemble genomes and identify functional genes from mixed populations. By capturing the collective genetic diversity, metagenomics enables comprehensive insights into microbial ecology, metabolic pathways, and environmental interactions.
Core Principles: How Single-Cell Genomics Works
Single-cell genomics isolates individual cells to analyze their complete genetic material, enabling high-resolution insights into cellular heterogeneity and gene expression profiles. Techniques such as microfluidics and fluorescence-activated cell sorting (FACS) facilitate the separation of single cells before whole genome amplification and sequencing. This contrasts with metagenomics, which sequences pooled DNA from entire microbial communities without isolating individual cells.
Data Generation and Sequencing Technologies
Metagenomics generates large-scale community DNA data using high-throughput sequencing platforms like Illumina and PacBio, enabling analysis of mixed microbial populations without culturing. Single-cell genomics isolates individual cells through microfluidics or FACS, followed by amplification methods like MDA to sequence their genomes with platforms such as Oxford Nanopore and 10x Genomics. These distinct data generation and sequencing technologies allow metagenomics to profile diversity and function at the ecosystem level, while single-cell genomics reveals heterogeneity and rare taxa within microbial communities.
Advantages of Metagenomics over Single-Cell Genomics
Metagenomics enables comprehensive analysis of entire microbial communities directly from environmental samples, capturing genetic diversity without the need for cell isolation. This approach offers higher throughput and cost-efficiency compared to the labor-intensive process of isolating and sequencing single cells. Moreover, metagenomics provides insights into microbial interactions and ecosystem functions that single-cell genomics may overlook due to its focus on individual cells.
Advantages of Single-Cell Genomics over Metagenomics
Single-cell genomics enables the analysis of individual cells, providing high-resolution insight into cellular heterogeneity and rare cell populations that metagenomics, which averages signals across microbial communities, cannot discern. It allows precise genome assembly and functional characterization from uncultivable microorganisms, overcoming the limitations of metagenomics related to mixed DNA samples and assembly biases. Single-cell genomics also facilitates tracking of genetic variations and evolutionary dynamics at the single-cell level, offering critical advantages for microbial ecology and pathogen detection.
Key Applications in Environmental and Clinical Research
Metagenomics enables comprehensive analysis of microbial communities in environmental samples, facilitating the study of biodiversity, ecosystem functions, and pollutant degradation. Single-cell genomics provides high-resolution insights into individual microbial cells, critical for identifying rare pathogens and understanding microbial heterogeneity in clinical infections. Both approaches complement each other in advancing diagnostic accuracy and ecological monitoring through precise genetic profiling.
Challenges and Limitations of Each Approach
Metagenomics faces challenges in assembling complex microbial communities due to high genomic diversity and the presence of closely related species, which complicates accurate taxonomic classification and functional annotation. Single-cell genomics encounters limitations including amplification bias, incomplete genome coverage, and technical difficulties in isolating individual cells from heterogeneous samples. Both methods require advanced bioinformatics tools to address data complexity and improve resolution for comprehensive microbial ecosystem analysis.
Integration of Metagenomics and Single-Cell Genomics
Integration of metagenomics and single-cell genomics enables comprehensive analysis of microbial communities by combining population-level genetic diversity with individual cell resolution. This approach enhances the identification of rare or unculturable species and uncovers metabolic pathways, improving insights into microbial ecology and functional potential. Advanced bioinformatics tools facilitate the fusion of datasets, driving breakthroughs in environmental microbiology and personalized medicine.
Future Trends and Innovations
Future trends in metagenomics emphasize high-throughput sequencing technologies coupled with advanced bioinformatics pipelines to unravel complex microbial communities with greater resolution. Single-cell genomics innovations focus on enhanced microfluidics and single-cell isolation techniques enabling precise genome assembly and functional analysis at an individual cell level. Integration of multi-omics data and AI-driven predictive models is poised to revolutionize microbial ecology and personalized medicine through deeper insights into cellular heterogeneity and dynamic interactions.
Metagenomics vs Single-cell genomics Infographic
 techiny.com
techiny.com