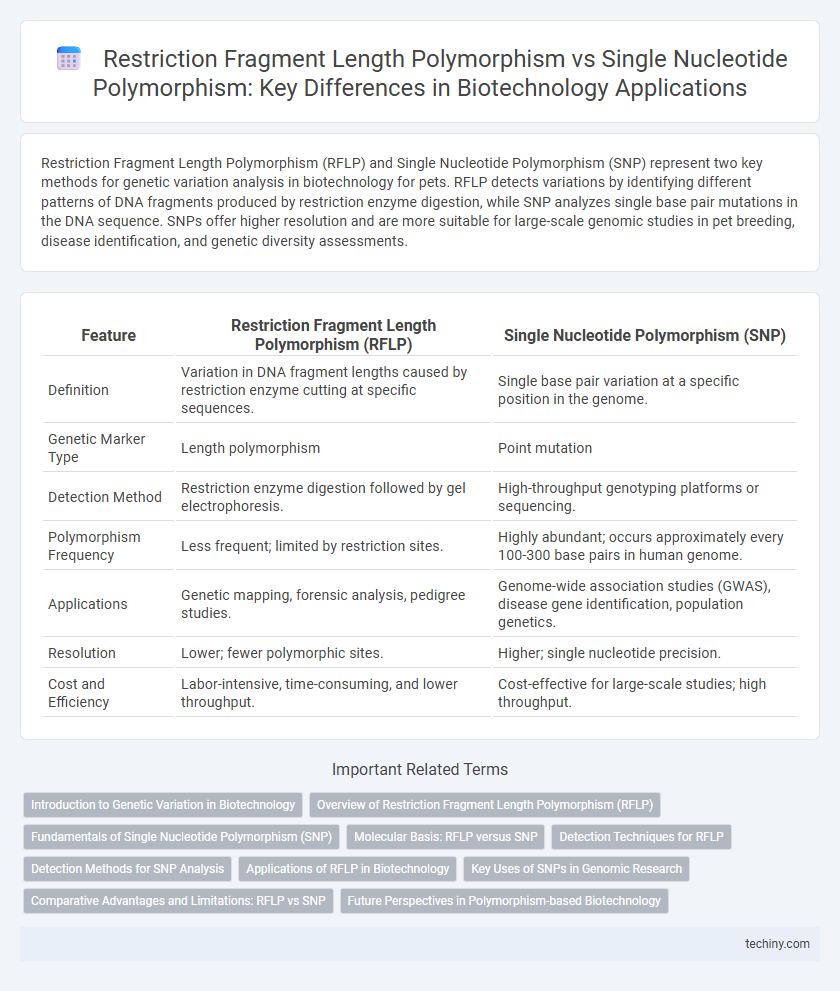
Restriction Fragment Length Polymorphism (RFLP) and Single Nucleotide Polymorphism (SNP) represent two key methods for genetic variation analysis in biotechnology for pets. RFLP detects variations by identifying different patterns of DNA fragments produced by restriction enzyme digestion, while SNP analyzes single base pair mutations in the DNA sequence. SNPs offer higher resolution and are more suitable for large-scale genomic studies in pet breeding, disease identification, and genetic diversity assessments.
Table of Comparison
| Feature | Restriction Fragment Length Polymorphism (RFLP) | Single Nucleotide Polymorphism (SNP) |
|---|---|---|
| Definition | Variation in DNA fragment lengths caused by restriction enzyme cutting at specific sequences. | Single base pair variation at a specific position in the genome. |
| Genetic Marker Type | Length polymorphism | Point mutation |
| Detection Method | Restriction enzyme digestion followed by gel electrophoresis. | High-throughput genotyping platforms or sequencing. |
| Polymorphism Frequency | Less frequent; limited by restriction sites. | Highly abundant; occurs approximately every 100-300 base pairs in human genome. |
| Applications | Genetic mapping, forensic analysis, pedigree studies. | Genome-wide association studies (GWAS), disease gene identification, population genetics. |
| Resolution | Lower; fewer polymorphic sites. | Higher; single nucleotide precision. |
| Cost and Efficiency | Labor-intensive, time-consuming, and lower throughput. | Cost-effective for large-scale studies; high throughput. |
Introduction to Genetic Variation in Biotechnology
Restriction Fragment Length Polymorphism (RFLP) and Single Nucleotide Polymorphism (SNP) are critical markers used to analyze genetic variation in biotechnology. RFLP detects variations in DNA fragment lengths caused by restriction enzyme cutting sites, while SNPs identify single base-pair changes in the genome, offering higher resolution for genetic diversity analysis. Understanding these markers enhances applications in genetic mapping, disease diagnosis, and plant and animal breeding programs.
Overview of Restriction Fragment Length Polymorphism (RFLP)
Restriction Fragment Length Polymorphism (RFLP) is a molecular technique used to detect variations in DNA sequences by identifying unique patterns created when specific restriction enzymes cut DNA at particular sites. This method analyzes differences in the length of DNA fragments resulting from polymorphisms, enabling genetic mapping, disease diagnosis, and forensic identification. RFLP is a robust tool for detecting large-scale genetic variation, contrasting with Single Nucleotide Polymorphism (SNP) analysis, which focuses on single base changes.
Fundamentals of Single Nucleotide Polymorphism (SNP)
Single Nucleotide Polymorphisms (SNPs) represent variations at a single base pair in the DNA sequence, making them the most abundant type of genetic variation in the genome. SNPs serve as crucial molecular markers in genetic mapping, disease association studies, and personalized medicine due to their stability and frequency across populations. Unlike Restriction Fragment Length Polymorphism (RFLP), which relies on variation in DNA fragment lengths after restriction enzyme digestion, SNP analysis provides higher resolution insights into genetic diversity and evolutionary relationships.
Molecular Basis: RFLP versus SNP
Restriction Fragment Length Polymorphism (RFLP) detects variations in DNA sequences by identifying differences in fragment lengths produced by specific restriction enzyme cuts, reflecting insertions, deletions, or mutations affecting enzyme recognition sites. Single Nucleotide Polymorphism (SNP) involves a single base pair variation at a specific position within the genome, representing the most common type of genetic variation in organisms. While RFLP assesses structural changes affecting restriction sites, SNP analysis focuses on single nucleotide substitutions, providing higher resolution for genetic mapping and association studies.
Detection Techniques for RFLP
Restriction Fragment Length Polymorphism (RFLP) detection techniques involve digesting DNA with specific restriction enzymes that recognize unique nucleotide sequences, resulting in fragment length variations analyzed through gel electrophoresis. Southern blotting is commonly used to transfer these fragments onto membranes for hybridization with labeled probes, enabling precise identification of polymorphic sites. Polymerase chain reaction (PCR) can also be integrated to amplify target regions before enzyme digestion, enhancing the sensitivity and specificity of RFLP analysis.
Detection Methods for SNP Analysis
Single Nucleotide Polymorphism (SNP) detection utilizes advanced methods such as TaqMan assays, SNP arrays, and next-generation sequencing (NGS) for high-throughput and precise genotyping. Restriction Fragment Length Polymorphism (RFLP) analysis relies on specific restriction enzymes to identify polymorphisms by fragment size differences through gel electrophoresis, but is less scalable compared to SNP genotyping techniques. Modern SNP detection methods offer higher sensitivity and efficiency, enabling large-scale genomic studies and personalized medicine applications in biotechnology.
Applications of RFLP in Biotechnology
Restriction Fragment Length Polymorphism (RFLP) is widely applied in biotechnology for genetic mapping, DNA fingerprinting, and detecting mutations linked to hereditary diseases. RFLP analysis enables precise identification of polymorphic regions within genomes, crucial for plant and animal breeding, forensic science, and microbial strain differentiation. Unlike Single Nucleotide Polymorphism (SNP) analysis, RFLP provides a robust approach for detecting larger-scale genetic variation and structural differences in DNA sequences.
Key Uses of SNPs in Genomic Research
Single Nucleotide Polymorphisms (SNPs) serve as crucial genetic markers in genomic research, enabling high-resolution mapping of disease loci and facilitating genome-wide association studies (GWAS) to identify genetic variants linked to complex traits. Unlike Restriction Fragment Length Polymorphisms (RFLPs), SNPs offer abundant and stable variation across populations, enhancing their utility in personalized medicine and pharmacogenomics. Their prevalence and distribution allow detailed analysis of population genetics, evolutionary biology, and the genetic basis of diseases.
Comparative Advantages and Limitations: RFLP vs SNP
Restriction Fragment Length Polymorphism (RFLP) offers high reliability and reproducibility in detecting genetic variations through enzyme digestion patterns, making it valuable for linkage mapping and forensic analysis despite being labor-intensive and requiring larger DNA amounts. Single Nucleotide Polymorphism (SNP) provides greater genome-wide coverage and high-throughput genotyping capabilities, enabling fine-scale association studies and personalized medicine, although SNP analysis can be limited by allele frequency and may miss larger structural variations. Both techniques complement each other, with RFLP excelling in precision for known polymorphisms and SNPs offering scalability and efficiency for diverse genetic research.
Future Perspectives in Polymorphism-based Biotechnology
Restriction Fragment Length Polymorphism (RFLP) and Single Nucleotide Polymorphism (SNP) serve as critical tools in polymorphism-based biotechnology, with SNPs offering higher genome-wide density and automation potential for future applications. Advances in high-throughput sequencing and bioinformatics are accelerating SNP genotyping efficiency, establishing it as a preferred marker for personalized medicine, crop improvement, and disease diagnostics. Integration of SNP analysis with CRISPR gene editing and AI-driven data interpretation promises to revolutionize precision breeding and therapeutic strategies by providing detailed genetic insights and scalable solutions.
Restriction Fragment Length Polymorphism vs Single Nucleotide Polymorphism Infographic
 techiny.com
techiny.com