Shotgun sequencing enables rapid analysis of complex genomes by randomly breaking DNA into small fragments and sequencing them simultaneously, making it ideal for large-scale projects. Sanger sequencing offers high accuracy through selective chain termination but is slower and better suited for sequencing smaller DNA regions or verifying shotgun results. Combining both techniques enhances the efficiency and precision of genetic analysis in biotechnology pet research.
Table of Comparison
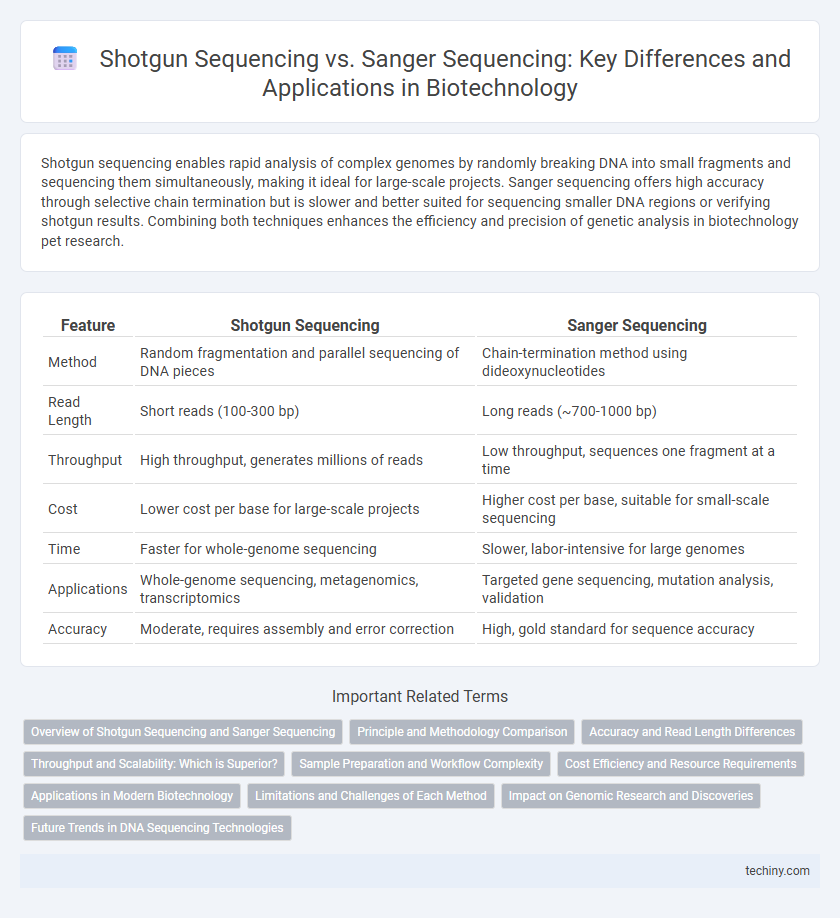
| Feature | Shotgun Sequencing | Sanger Sequencing |
|---|---|---|
| Method | Random fragmentation and parallel sequencing of DNA pieces | Chain-termination method using dideoxynucleotides |
| Read Length | Short reads (100-300 bp) | Long reads (~700-1000 bp) |
| Throughput | High throughput, generates millions of reads | Low throughput, sequences one fragment at a time |
| Cost | Lower cost per base for large-scale projects | Higher cost per base, suitable for small-scale sequencing |
| Time | Faster for whole-genome sequencing | Slower, labor-intensive for large genomes |
| Applications | Whole-genome sequencing, metagenomics, transcriptomics | Targeted gene sequencing, mutation analysis, validation |
| Accuracy | Moderate, requires assembly and error correction | High, gold standard for sequence accuracy |
Overview of Shotgun Sequencing and Sanger Sequencing
Shotgun sequencing involves randomly breaking DNA into numerous small fragments, sequencing them individually, and then using computational methods to assemble the sequences back into a complete genome, making it efficient for large-scale genome projects. Sanger sequencing, also known as chain-termination sequencing, utilizes selective incorporation of dideoxynucleotides during DNA replication to generate DNA fragments of varying lengths that are then separated by electrophoresis, providing highly accurate and longer read lengths ideal for smaller DNA regions. Both methods serve critical roles in genomics, with shotgun sequencing favoring high-throughput genome assembly and Sanger sequencing excelling in targeted sequencing tasks requiring precision.
Principle and Methodology Comparison
Shotgun sequencing employs random fragmentation of DNA followed by sequencing of numerous overlapping fragments, allowing for rapid assembly of entire genomes using computational algorithms. In contrast, Sanger sequencing relies on selective incorporation of chain-terminating dideoxynucleotides during DNA synthesis, producing high-accuracy reads of shorter DNA fragments. Shotgun sequencing is advantageous for large-scale genomic projects due to its high throughput and speed, while Sanger sequencing remains the gold standard for precise sequencing of smaller DNA regions and validation purposes.
Accuracy and Read Length Differences
Shotgun sequencing offers high accuracy by generating numerous overlapping short reads, enabling reliable genome assembly despite shorter individual read lengths typically around 150-300 base pairs. In contrast, Sanger sequencing produces longer reads averaging 800-1000 base pairs with very high per-base accuracy, making it ideal for sequencing smaller genomic regions or validating shotgun results. The trade-off between read length and throughput defines their usage, with shotgun sequencing dominating large-scale whole-genome projects and Sanger sequencing preferred for targeted, high-fidelity sequence verification.
Throughput and Scalability: Which is Superior?
Shotgun sequencing offers significantly higher throughput and scalability compared to Sanger sequencing by fragmenting DNA into millions of small pieces and sequencing them simultaneously using high-throughput next-generation platforms. In contrast, Sanger sequencing processes DNA fragments sequentially, limiting its throughput to lower numbers of base pairs per run and making it less scalable for large genomes. Therefore, Shotgun sequencing is superior for large-scale genomic projects requiring rapid and extensive data generation.
Sample Preparation and Workflow Complexity
Shotgun sequencing requires fragmenting the entire genome into small, random pieces, demanding high-quality DNA extraction and extensive library preparation to ensure even coverage. Sanger sequencing involves targeted amplification of specific DNA regions using primers, resulting in simpler sample preparation but limited throughput. The workflow of shotgun sequencing is more complex due to massive parallel processing and computational assembly, whereas Sanger sequencing follows a more straightforward, linear process ideal for lower-volume, high-accuracy sequencing.
Cost Efficiency and Resource Requirements
Shotgun sequencing offers higher cost efficiency compared to Sanger sequencing due to its ability to process large genomes rapidly using parallelized methods. Sanger sequencing requires more resources, including expensive reagents and labor-intensive sample preparation, making it less practical for large-scale projects. Despite higher initial investment in computational infrastructure, shotgun sequencing reduces overall expenses by minimizing time and manual intervention.
Applications in Modern Biotechnology
Shotgun sequencing enables high-throughput decoding of entire genomes, making it indispensable for large-scale projects like metagenomics and de novo genome assembly in modern biotechnology. Sanger sequencing remains the gold standard for validating specific gene sequences, performing mutation analysis, and sequencing small DNA fragments with high accuracy. Together, these techniques complement each other by balancing rapid, comprehensive genomic data acquisition with precise, targeted genetic analysis.
Limitations and Challenges of Each Method
Shotgun sequencing faces limitations such as difficulty in assembling highly repetitive genomic regions and higher error rates due to short read lengths, which complicate accurate genome reconstruction. Sanger sequencing, while providing longer and more accurate reads, is challenged by lower throughput and higher cost, making it inefficient for large-scale genome projects. Both methods require robust computational tools for data analysis, yet Sanger's scalability constraints hinder its application in modern high-throughput sequencing demands.
Impact on Genomic Research and Discoveries
Shotgun sequencing revolutionized genomic research by enabling rapid, large-scale DNA analysis, dramatically accelerating genome assembly and discovery of complex genetic variations. In contrast, Sanger sequencing, with its high accuracy but lower throughput, remains invaluable for validating specific gene sequences and small-scale projects. The combination of both methods has enhanced our understanding of genetic diversity, disease mechanisms, and personalized medicine.
Future Trends in DNA Sequencing Technologies
Shotgun sequencing enables rapid, large-scale genome analysis by randomly fragmenting DNA and assembling sequences via computational methods, contrasting with the targeted, high-accuracy approach of Sanger sequencing. Future trends in DNA sequencing emphasize enhanced throughput, cost reduction, and real-time data analysis, with third-generation technologies like nanopore and single-molecule sequencing advancing precision and read length beyond traditional methods. Integration of AI-driven bioinformatics tools promises to accelerate genomic interpretation and personalized medicine applications.
**Shotgun sequencing vs Sanger sequencing** Infographic
 techiny.com
techiny.com