Shotgun sequencing rapidly decodes entire genomes by randomly breaking DNA into small fragments, allowing high-throughput analysis ideal for complex organisms. Clone-by-clone sequencing involves mapping and sequencing individual cloned DNA segments, offering higher accuracy and easier assembly for large genomes but requires more time and resources. Choosing between these methods depends on project scale, accuracy needs, and available computational tools in biotechnology research.
Table of Comparison
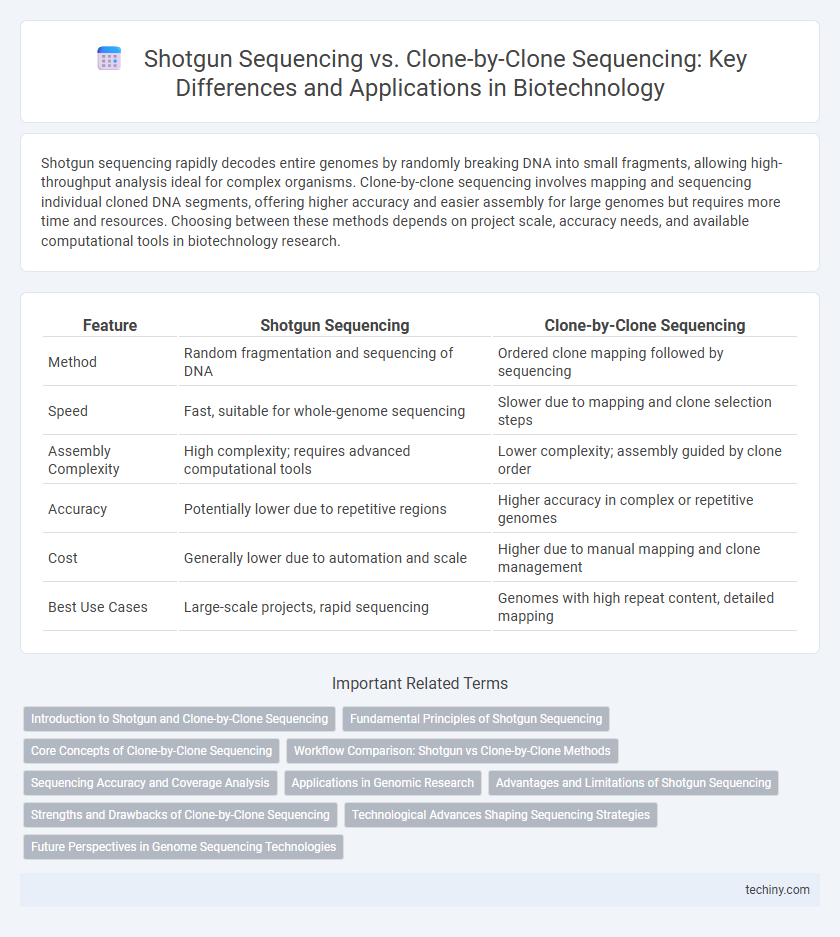
| Feature | Shotgun Sequencing | Clone-by-Clone Sequencing |
|---|---|---|
| Method | Random fragmentation and sequencing of DNA | Ordered clone mapping followed by sequencing |
| Speed | Fast, suitable for whole-genome sequencing | Slower due to mapping and clone selection steps |
| Assembly Complexity | High complexity; requires advanced computational tools | Lower complexity; assembly guided by clone order |
| Accuracy | Potentially lower due to repetitive regions | Higher accuracy in complex or repetitive genomes |
| Cost | Generally lower due to automation and scale | Higher due to manual mapping and clone management |
| Best Use Cases | Large-scale projects, rapid sequencing | Genomes with high repeat content, detailed mapping |
Introduction to Shotgun and Clone-by-Clone Sequencing
Shotgun sequencing involves randomly breaking DNA into numerous small fragments for parallel sequencing, offering rapid genome analysis with high-throughput efficiency. Clone-by-clone sequencing requires dividing the genome into large DNA clones, mapping each clone, and sequencing them individually, providing a more organized but time-consuming approach. The choice between these methods depends on factors like genome complexity, available resources, and desired accuracy.
Fundamental Principles of Shotgun Sequencing
Shotgun sequencing involves randomly breaking DNA into numerous small fragments, sequencing them independently, and using computational algorithms to assemble overlapping sequences into a complete genome. This method contrasts with clone-by-clone sequencing, which sequences large cloned DNA segments in an ordered manner before assembly. The fundamental principle of shotgun sequencing lies in its reliance on high-throughput sequencing technologies combined with bioinformatics to reconstruct complex genomes rapidly and cost-effectively.
Core Concepts of Clone-by-Clone Sequencing
Clone-by-clone sequencing divides the genome into large, mapped clones that are individually sequenced to ensure accurate assembly and reduce errors in repetitive regions. Each clone is sequenced separately, allowing for easier reconstruction of complex genomes by creating a physical map before sequencing. This hierarchical approach contrasts with shotgun sequencing's random fragmentation, providing higher confidence in genome structure and reducing ambiguity.
Workflow Comparison: Shotgun vs Clone-by-Clone Methods
Shotgun sequencing involves randomly breaking the genome into small fragments, sequencing them, and then using computational methods to assemble overlapping sequences into a continuous genome, enabling faster and high-throughput analysis. Clone-by-clone sequencing requires the division of the genome into large, ordered fragments cloned into vectors, which are mapped and then individually sequenced, ensuring high accuracy and easier assembly of repetitive regions. The shotgun method offers speed and scalability, while clone-by-clone sequencing provides greater structural clarity, especially for complex genomes.
Sequencing Accuracy and Coverage Analysis
Shotgun sequencing offers extensive coverage by randomly breaking DNA into small fragments, enabling rapid analysis but potentially introducing gaps or errors due to overlapping regions. Clone-by-clone sequencing provides higher accuracy through ordered, overlapping clone maps, ensuring thorough coverage with precise alignment, though it is more time-consuming and labor-intensive. Accuracy in clone-by-clone sequencing benefits from reduced ambiguity in assembly, whereas shotgun sequencing relies heavily on computational algorithms to resolve overlaps and correct sequencing errors.
Applications in Genomic Research
Shotgun sequencing enables rapid whole-genome analysis by randomly fragmenting DNA and assembling sequences computationally, making it ideal for high-throughput genomic projects such as complex genome sequencing and metagenomics. Clone-by-clone sequencing, relying on hierarchical mapping of large DNA fragments before sequencing, provides precise structural information critical for detailed annotation and verification of genomic regions in model organisms. Both methods complement each other in genomic research, with shotgun sequencing accelerating coverage and clone-by-clone ensuring accuracy in complex or repetitive sequences.
Advantages and Limitations of Shotgun Sequencing
Shotgun sequencing offers rapid genome analysis by randomly breaking DNA into small fragments for parallel sequencing, significantly reducing time and cost compared to clone-by-clone sequencing. Its major advantage lies in high throughput and efficiency, enabling comprehensive coverage without requiring a physical map of the genome. Limitations include challenges in assembling repetitive regions and potential gaps due to uneven fragment coverage, which may complicate complete genome reconstruction.
Strengths and Drawbacks of Clone-by-Clone Sequencing
Clone-by-clone sequencing offers high accuracy and reliability by mapping large DNA fragments before sequencing, minimizing assembly errors and simplifying genome assembly of complex regions. This method is particularly effective for large genomes with repetitive sequences, providing a structured approach that reduces computational challenges compared to shotgun sequencing. However, clone-by-clone sequencing is time-consuming, labor-intensive, and costlier due to the necessity of creating and screening physical maps prior to sequencing.
Technological Advances Shaping Sequencing Strategies
Shotgun sequencing leverages high-throughput automation and parallel processing to rapidly generate overlapping DNA fragments, enabling faster genome assembly compared to clone-by-clone sequencing's methodical, hierarchical approach. Advances in next-generation sequencing (NGS) technologies such as Illumina and nanopore platforms have enhanced the efficiency and accuracy of shotgun strategies, reducing costs and turnaround times. Clone-by-clone sequencing remains valuable for complex regions requiring precise mapping, but integration with computational algorithms and long-read sequencing continues to shift the balance towards shotgun methods in genomic research.
Future Perspectives in Genome Sequencing Technologies
Shotgun sequencing offers rapid, high-throughput genome analysis by fragmenting DNA and sequencing randomly, enabling efficient handling of complex genomes, while clone-by-clone sequencing provides high accuracy through ordered mapping but is more time-consuming. Advances in long-read technologies and AI-driven assembly algorithms are expected to enhance shotgun sequencing accuracy, potentially overcoming its limitations in repetitive regions. Future genome sequencing technologies will likely integrate hybrid approaches, combining the speed of shotgun methods with the precision of clone-by-clone mapping to achieve comprehensive, cost-effective genomic insights.
Shotgun Sequencing vs Clone-by-Clone Sequencing Infographic
 techiny.com
techiny.com