SNP genotyping focuses on identifying single nucleotide polymorphisms that provide detailed insights into genetic variation and disease susceptibility in pets. CNV analysis detects larger structural changes such as deletions or duplications of DNA segments, which can significantly impact gene function and contribute to complex traits or disorders. Both approaches complement each other by offering a comprehensive view of genetic diversity and health risks in biotechnology applications for pets.
Table of Comparison
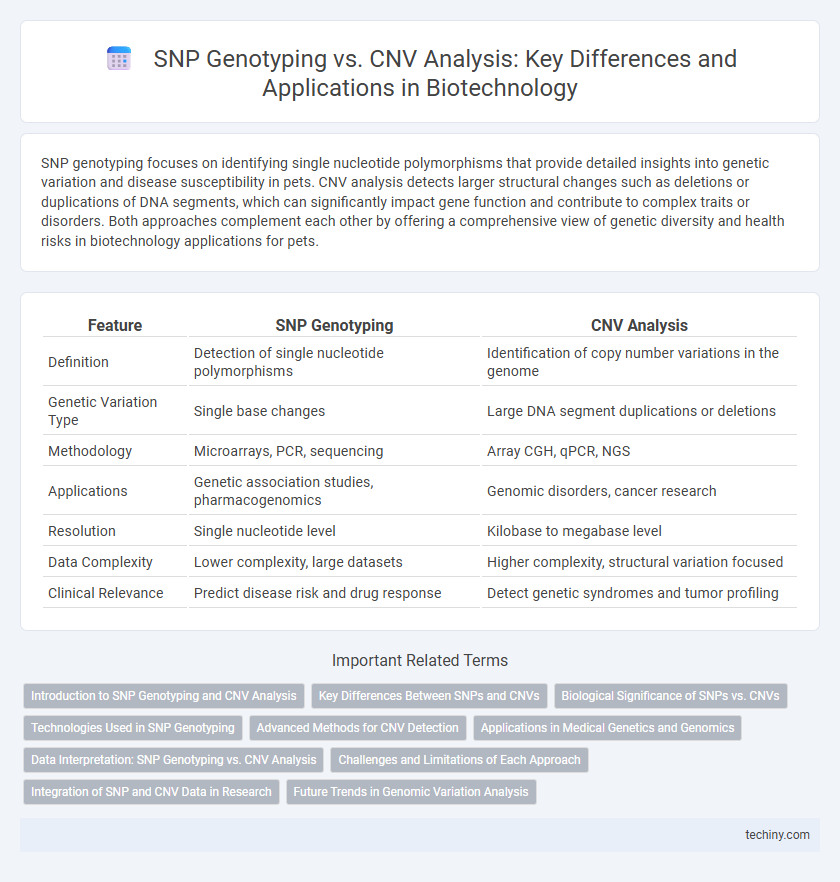
| Feature | SNP Genotyping | CNV Analysis |
|---|---|---|
| Definition | Detection of single nucleotide polymorphisms | Identification of copy number variations in the genome |
| Genetic Variation Type | Single base changes | Large DNA segment duplications or deletions |
| Methodology | Microarrays, PCR, sequencing | Array CGH, qPCR, NGS |
| Applications | Genetic association studies, pharmacogenomics | Genomic disorders, cancer research |
| Resolution | Single nucleotide level | Kilobase to megabase level |
| Data Complexity | Lower complexity, large datasets | Higher complexity, structural variation focused |
| Clinical Relevance | Predict disease risk and drug response | Detect genetic syndromes and tumor profiling |
Introduction to SNP Genotyping and CNV Analysis
SNP genotyping identifies single nucleotide polymorphisms, enabling detailed analysis of genetic variation at specific loci, whereas CNV analysis detects copy number variations involving large DNA segments, revealing structural genomic alterations. Both techniques utilize high-throughput sequencing or microarray platforms to provide complementary insights into genetic diversity and disease susceptibility. Accurate detection of SNPs and CNVs is crucial in personalized medicine, pharmacogenomics, and complex trait mapping.
Key Differences Between SNPs and CNVs
SNP genotyping identifies single nucleotide variations at specific genomic positions, providing detailed insights into genetic diversity and association with complex traits. CNV analysis detects larger genomic segments that vary in copy number, impacting gene dosage and structural variation within the genome. Unlike SNPs, which represent point mutations, CNVs encompass deletions, duplications, and insertions that significantly affect genomic architecture and phenotypic variability.
Biological Significance of SNPs vs. CNVs
SNP genotyping and CNV analysis both reveal critical genetic variations influencing phenotypic diversity and disease susceptibility. Single nucleotide polymorphisms (SNPs) represent the most abundant form of genetic variation, affecting gene function and regulation at a granular level, often linked to complex traits and pharmacogenomics. Copy number variations (CNVs) encompass larger genomic segments affecting gene dosage and structural genomic integrity, frequently associated with developmental disorders, cancer, and adaptive evolution.
Technologies Used in SNP Genotyping
SNP genotyping relies primarily on technologies such as microarrays, next-generation sequencing (NGS), and allele-specific PCR, enabling high-throughput and precise detection of single nucleotide polymorphisms across genomes. Microarray platforms, like Illumina Infinium and Affymetrix Axiom, offer extensive SNP coverage for genome-wide association studies, while NGS provides sequence-level resolution facilitating novel variant discovery. Allele-specific PCR techniques complement these methods by targeting known SNPs with high specificity and cost-effectiveness in validation studies.
Advanced Methods for CNV Detection
Advanced methods for CNV detection in biotechnology leverage high-resolution techniques such as next-generation sequencing (NGS) and array comparative genomic hybridization (aCGH) for precise identification of copy number variations. These techniques provide superior sensitivity and specificity compared to traditional SNP genotyping arrays, enabling the detection of both large and submicroscopic CNVs with greater accuracy. Integration of machine learning algorithms further enhances CNV interpretation, facilitating comprehensive genomic profiling in clinical and research applications.
Applications in Medical Genetics and Genomics
SNP genotyping enables precise identification of single nucleotide polymorphisms, critical for associating genetic variants with complex diseases and personalized medicine strategies. CNV analysis detects larger structural variations in the genome, such as duplications or deletions, which are key in diagnosing developmental disorders, cancer genomics, and neuropsychiatric conditions. Integrating both methods enhances comprehensive genomic profiling, improving diagnostic accuracy and targeted therapeutic interventions in medical genetics.
Data Interpretation: SNP Genotyping vs. CNV Analysis
SNP genotyping provides precise detection of single nucleotide variations, enabling detailed mapping of genetic diversity and disease association studies with high-resolution allelic discrimination. CNV analysis identifies larger structural variations such as deletions and duplications across the genome, offering insights into gene dosage effects and their impact on phenotypic traits and complex diseases. Data interpretation in SNP genotyping focuses on single-base precision and allele frequency, while CNV analysis requires integrating signal intensity and copy number thresholds to assess genomic structural variation.
Challenges and Limitations of Each Approach
SNP genotyping faces challenges such as limited detection of rare variants and difficulties in distinguishing closely linked polymorphisms, impacting its resolution in complex genomic regions. CNV analysis struggles with accurately quantifying copy number changes in highly repetitive sequences and heterogeneous samples, leading to potential false positives or negatives. Both approaches require high-quality DNA and advanced computational tools to mitigate artifacts and ensure reliable interpretation of genetic variation.
Integration of SNP and CNV Data in Research
Integrating SNP genotyping and CNV analysis enhances genomic research by providing a comprehensive view of genetic variation that impacts phenotypic traits and disease susceptibility. Combining single nucleotide polymorphisms (SNPs) with copy number variations (CNVs) allows for more precise identification of genetic markers and improves the accuracy of genome-wide association studies (GWAS). Advanced bioinformatics tools enable the simultaneous analysis of SNP and CNV data, facilitating the discovery of novel genetic interactions and contributing to personalized medicine and targeted therapeutics.
Future Trends in Genomic Variation Analysis
Future trends in genomic variation analysis emphasize the integration of SNP genotyping and CNV analysis through high-throughput sequencing technologies to enhance precision medicine. Advances in bioinformatics tools are enabling more comprehensive detection of both single nucleotide polymorphisms (SNPs) and copy number variations (CNVs), improving genetic disease diagnosis and treatment customization. Machine learning algorithms are increasingly applied to interpret complex genomic data, driving the identification of novel biomarkers and facilitating large-scale population genomics studies.
SNP Genotyping vs CNV Analysis Infographic
 techiny.com
techiny.com