Site-directed mutagenesis enables precise alterations at specific DNA locations, allowing targeted study of gene function in biotechnology pet research. Random mutagenesis generates diverse genetic variations without predetermined sites, facilitating the discovery of novel traits and enhanced phenotypes. Both techniques play crucial roles in advancing genetic engineering and improving pet health through tailored molecular interventions.
Table of Comparison
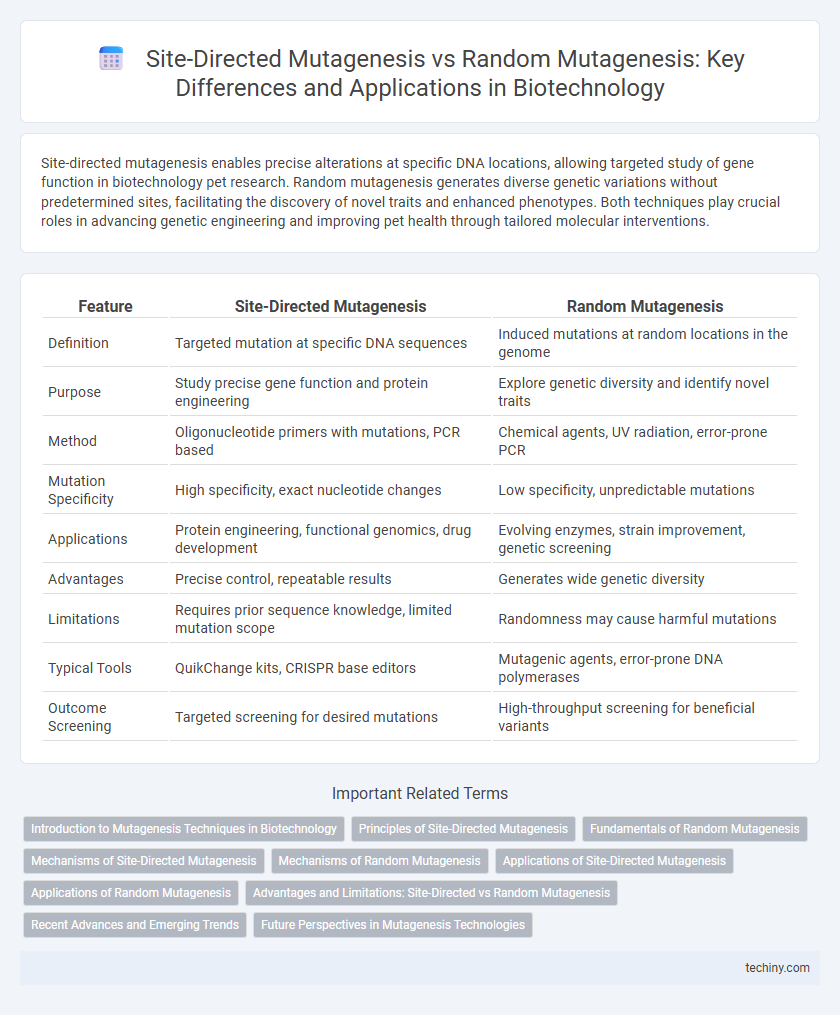
| Feature | Site-Directed Mutagenesis | Random Mutagenesis |
|---|---|---|
| Definition | Targeted mutation at specific DNA sequences | Induced mutations at random locations in the genome |
| Purpose | Study precise gene function and protein engineering | Explore genetic diversity and identify novel traits |
| Method | Oligonucleotide primers with mutations, PCR based | Chemical agents, UV radiation, error-prone PCR |
| Mutation Specificity | High specificity, exact nucleotide changes | Low specificity, unpredictable mutations |
| Applications | Protein engineering, functional genomics, drug development | Evolving enzymes, strain improvement, genetic screening |
| Advantages | Precise control, repeatable results | Generates wide genetic diversity |
| Limitations | Requires prior sequence knowledge, limited mutation scope | Randomness may cause harmful mutations |
| Typical Tools | QuikChange kits, CRISPR base editors | Mutagenic agents, error-prone DNA polymerases |
| Outcome Screening | Targeted screening for desired mutations | High-throughput screening for beneficial variants |
Introduction to Mutagenesis Techniques in Biotechnology
Site-directed mutagenesis enables precise alterations of specific DNA sequences to study gene function or engineer proteins, offering targeted and predictable genetic modifications. In contrast, random mutagenesis generates diverse genetic variants across the genome or within specific regions, facilitating the discovery of novel phenotypes through high-throughput screening. These mutagenesis techniques are fundamental in biotechnology for protein engineering, functional genomics, and developing genetically modified organisms.
Principles of Site-Directed Mutagenesis
Site-directed mutagenesis employs precise alteration of specific DNA sequences using strategies such as primer extension or PCR-based methods, enabling targeted amino acid substitutions to study protein function. This technique relies on synthetic oligonucleotides containing the desired mutation to hybridize with template DNA, followed by enzymatic synthesis and selective degradation of parental strands. The method allows for controlled, reproducible genetic modifications compared to random mutagenesis, which generates diverse mutations without specificity.
Fundamentals of Random Mutagenesis
Random mutagenesis introduces genetic variability by inducing mutations throughout the entire genome or target gene without specific site restriction, typically using chemical mutagens, UV radiation, or error-prone PCR techniques. This approach enables the exploration of a wide range of mutations, facilitating the discovery of novel phenotypes and protein functions that may not be predicted by rational design. It contrasts with site-directed mutagenesis, which targets precise nucleotide changes, providing a broader spectrum of genetic diversity but less control over mutation locations.
Mechanisms of Site-Directed Mutagenesis
Site-directed mutagenesis involves precise alteration of specific nucleotide sequences within a gene using techniques like oligonucleotide-directed mutagenesis, PCR-based methods, or CRISPR-Cas systems. This mechanism enables targeted substitution, insertion, or deletion by introducing synthetic DNA primers that anneal to the template strand and guide DNA polymerase to incorporate desired mutations during replication. The high specificity of site-directed mutagenesis makes it invaluable for investigating protein function, enzyme activity, and gene regulation in biotechnology and molecular biology research.
Mechanisms of Random Mutagenesis
Random mutagenesis operates through mechanisms that introduce spontaneous or induced mutations across the genome, often utilizing chemical mutagens, radiation, or error-prone PCR to generate a diverse pool of genetic variants. These mutations occur without target specificity, affecting multiple loci and enabling broad phenotypic exploration essential for directed evolution studies. This contrasts with site-directed mutagenesis, which employs precise nucleotide substitutions guided by designed primers to alter specific gene sites.
Applications of Site-Directed Mutagenesis
Site-directed mutagenesis enables precise alteration of DNA sequences to study gene function, protein engineering, and drug development, making it invaluable for creating specific amino acid substitutions in enzymes and therapeutic proteins. This technique allows targeted modification of genetic sequences to investigate the effects of mutations on protein activity, stability, and interaction with ligands, accelerating the design of enhanced biomolecules. Site-directed mutagenesis is widely applied in functional genomics, antibody optimization, and metabolic pathway engineering to improve industrial biocatalysts and develop novel biopharmaceuticals.
Applications of Random Mutagenesis
Random mutagenesis generates genetic diversity by introducing mutations at random locations within a gene or genome, enhancing the development of proteins with novel or improved functions in biotechnology. It is extensively applied in enzyme engineering, strain improvement for industrial fermentation, and the discovery of variants with increased stability or activity. This approach accelerates adaptive evolution studies and complements rational design by exploring sequence space beyond known functional constraints.
Advantages and Limitations: Site-Directed vs Random Mutagenesis
Site-directed mutagenesis enables precise and targeted alterations in a gene sequence, leading to specific amino acid substitutions that facilitate detailed functional studies of proteins. This method's limitation lies in its requirement for prior knowledge of gene structure, making it less effective for discovering novel mutations compared to random mutagenesis. Random mutagenesis generates a diverse pool of variants without predetermined sites, advantageous for exploring unknown gene functions but often necessitating extensive screening to identify beneficial mutations.
Recent Advances and Emerging Trends
Site-directed mutagenesis enables precise nucleotide alterations in specific gene sequences, facilitating targeted protein engineering and functional studies, while random mutagenesis generates diverse genetic variants through error-prone PCR or chemical mutagens, accelerating the discovery of novel phenotypes. Recent advances include CRISPR-Cas systems enhancing site-directed mutagenesis accuracy and high-throughput sequencing improving the identification of beneficial random mutations. Emerging trends focus on integrating machine learning algorithms with mutagenesis data to predict mutation impacts and optimize enzyme design in synthetic biology applications.
Future Perspectives in Mutagenesis Technologies
Site-directed mutagenesis enables precise genetic modifications, fostering advancements in gene therapy and protein engineering with targeted, predictable outcomes. Random mutagenesis accelerates the exploration of genetic diversity, facilitating the discovery of novel traits and pathways through high-throughput screening methods. Emerging technologies such as CRISPR-based base editing and machine learning algorithms are poised to enhance the efficiency, accuracy, and scalability of both mutagenesis approaches, revolutionizing synthetic biology and personalized medicine.
Site-Directed Mutagenesis vs Random Mutagenesis Infographic
 techiny.com
techiny.com