In situ hybridization (ISH) and fluorescence in situ hybridization (FISH) are essential techniques used in biotechnology for detecting specific nucleic acid sequences within cells. ISH utilizes labeled probes to visualize gene expression patterns, while FISH employs fluorescently tagged probes for enhanced sensitivity and spatial resolution of chromosomal abnormalities. Both methods enable precise localization of genetic material, with FISH offering more detailed three-dimensional imaging useful in complex genetic analyses.
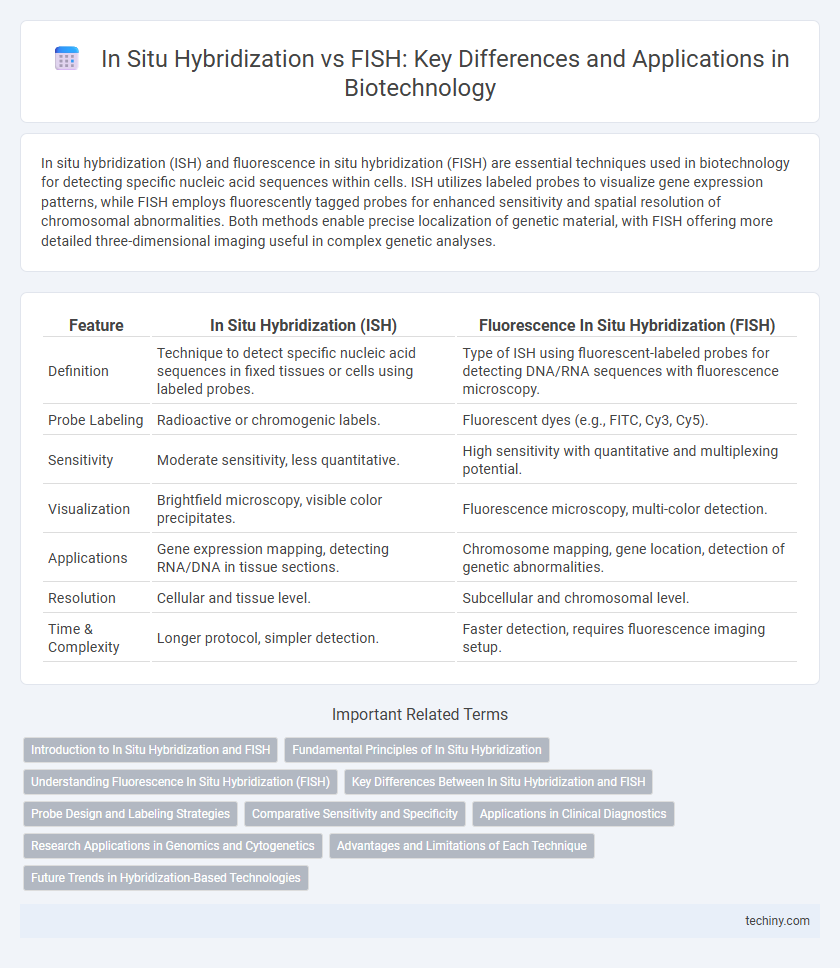
Table of Comparison
| Feature | In Situ Hybridization (ISH) | Fluorescence In Situ Hybridization (FISH) |
|---|---|---|
| Definition | Technique to detect specific nucleic acid sequences in fixed tissues or cells using labeled probes. | Type of ISH using fluorescent-labeled probes for detecting DNA/RNA sequences with fluorescence microscopy. |
| Probe Labeling | Radioactive or chromogenic labels. | Fluorescent dyes (e.g., FITC, Cy3, Cy5). |
| Sensitivity | Moderate sensitivity, less quantitative. | High sensitivity with quantitative and multiplexing potential. |
| Visualization | Brightfield microscopy, visible color precipitates. | Fluorescence microscopy, multi-color detection. |
| Applications | Gene expression mapping, detecting RNA/DNA in tissue sections. | Chromosome mapping, gene location, detection of genetic abnormalities. |
| Resolution | Cellular and tissue level. | Subcellular and chromosomal level. |
| Time & Complexity | Longer protocol, simpler detection. | Faster detection, requires fluorescence imaging setup. |
Introduction to In Situ Hybridization and FISH
In situ hybridization (ISH) is a molecular technique used to detect specific nucleic acid sequences within fixed tissues or cells, enabling spatial localization of gene expression. Fluorescence in situ hybridization (FISH) is an advanced form of ISH that employs fluorescent probes for precise visualization of DNA or RNA sequences under a fluorescence microscope. Both ISH and FISH are essential tools in biotechnology for studying genetic abnormalities, gene mapping, and cellular transcriptional activity with high specificity and resolution.
Fundamental Principles of In Situ Hybridization
In situ hybridization (ISH) relies on the hybridization of a labeled nucleic acid probe to a specific complementary DNA or RNA sequence within fixed cells or tissue sections, enabling precise localization of gene expression. Fluorescence in situ hybridization (FISH) is a subtype of ISH that utilizes fluorescently labeled probes for visualization under a fluorescence microscope, enhancing resolution and multiplexing capabilities. Fundamental principles of ISH include probe design specificity, nucleic acid target accessibility, hybridization conditions, and stringent washing steps to ensure accurate and specific detection of target sequences.
Understanding Fluorescence In Situ Hybridization (FISH)
Fluorescence In Situ Hybridization (FISH) utilizes fluorescent probes to bind specific DNA sequences on chromosomes, enabling precise visualization of genetic abnormalities directly within cells. Compared to traditional in situ hybridization, FISH offers higher sensitivity and spatial resolution, facilitating the detection of chromosomal translocations, gene amplifications, and deletions in clinical diagnostics and research. The technique's ability to provide real-time localization of target sequences enhances genetic analysis in oncology, prenatal screening, and microbial identification.
Key Differences Between In Situ Hybridization and FISH
In situ hybridization (ISH) detects specific nucleic acid sequences within fixed tissues or cells using labeled DNA or RNA probes, enabling spatial localization of gene expression. Fluorescence in situ hybridization (FISH) is a subtype of ISH that employs fluorescently labeled probes for high-resolution visualization of chromosomal abnormalities and gene mapping under a fluorescence microscope. Key differences include FISH's use of fluorescence for precise detection and its application predominantly in cytogenetics, whereas ISH encompasses broader probe types and is applied for general mRNA or DNA localization studies.
Probe Design and Labeling Strategies
In situ hybridization (ISH) and fluorescence in situ hybridization (FISH) both rely on careful probe design to ensure target specificity and signal clarity; ISH probes are typically longer RNA or DNA sequences that hybridize to complementary nucleic acid targets, while FISH probes are often shorter, fluorescently labeled oligonucleotides optimized for rapid hybridization and high signal-to-noise ratios. Labeling strategies for ISH commonly involve enzymatic incorporation of digoxigenin or biotin, followed by chromogenic detection, whereas FISH utilizes directly conjugated fluorescent dyes such as Alexa Fluor or Cy dyes for multiplexed and quantitative imaging. The choice of probe length, labeling chemistry, and hybridization conditions critically impacts detection sensitivity, spatial resolution, and the ability to discriminate single-nucleotide polymorphisms in complex biological samples.
Comparative Sensitivity and Specificity
In situ hybridization (ISH) and fluorescence in situ hybridization (FISH) are both powerful techniques for detecting specific nucleic acid sequences within cells, yet FISH generally offers higher sensitivity and specificity due to its use of fluorescent probes that provide clearer signal-to-noise ratios. Comparative analyses reveal that FISH allows for more precise localization of target sequences at the subcellular level, reducing background interference common in chromogenic ISH. Studies in clinical diagnostics and research confirm that FISH's enhanced fluorescence detection results in improved accuracy for identifying genetic abnormalities and pathogen presence compared to traditional ISH methods.
Applications in Clinical Diagnostics
In situ hybridization (ISH) and fluorescence in situ hybridization (FISH) are pivotal techniques in clinical diagnostics for detecting specific nucleic acid sequences within tissue samples. FISH offers higher sensitivity and multiplexing capabilities, enabling precise identification of chromosomal abnormalities, gene amplifications, and translocations associated with cancer, genetic disorders, and infectious diseases. ISH is widely utilized for analyzing gene expression patterns at the cellular level, providing valuable insights into tumor pathology and aiding in personalized treatment strategies.
Research Applications in Genomics and Cytogenetics
In situ hybridization (ISH) enables precise localization of specific nucleic acid sequences within tissue sections, facilitating gene expression analysis in developmental biology and cancer research. Fluorescence in situ hybridization (FISH) enhances cytogenetic studies by using fluorescent probes to detect chromosomal abnormalities, gene amplifications, and translocations with high sensitivity. Both techniques are pivotal in genomics for mapping genetic sequences and diagnosing genetic disorders, with FISH offering multiplexing capabilities for complex chromosomal investigations.
Advantages and Limitations of Each Technique
In situ hybridization (ISH) offers a versatile approach for detecting specific nucleic acid sequences within fixed tissues, enabling precise localization of gene expression patterns at cellular resolution with relatively straightforward protocols. Fluorescence in situ hybridization (FISH) enhances sensitivity and multiplexing capacity by employing fluorescent probes, allowing simultaneous visualization of multiple targets and enabling detailed chromosomal mapping, yet it requires specialized fluorescence microscopy and can suffer from photobleaching. While ISH provides a cost-effective and accessible method suitable for various sample types, its lower sensitivity and limited multiplexing restrict its use compared to FISH, which, despite higher technical demands and costs, delivers superior spatial resolution and quantitative analysis in complex genomic studies.
Future Trends in Hybridization-Based Technologies
Advancements in hybridization-based technologies are steering towards combining In situ hybridization (ISH) with high-throughput sequencing and multiplexed fluorescence, enhancing spatial resolution and molecular profiling simultaneously. Future trends emphasize automation and AI-driven image analysis to improve accuracy and speed in diagnostic applications, particularly in oncology and personalized medicine. Integration of novel probes and nanotechnology promises greater sensitivity and the ability to detect multiple targets within single cells or tissues.
In situ hybridization vs FISH Infographic
 techiny.com
techiny.com