Bayesian Networks represent probabilistic relationships through directed acyclic graphs, capturing causal dependencies among variables. Markov Random Fields model undirected dependencies, emphasizing local interactions and conditional independencies in a graph structure. Both frameworks enable efficient probabilistic inference but differ in how they represent and reason about uncertainty in complex systems.
Table of Comparison
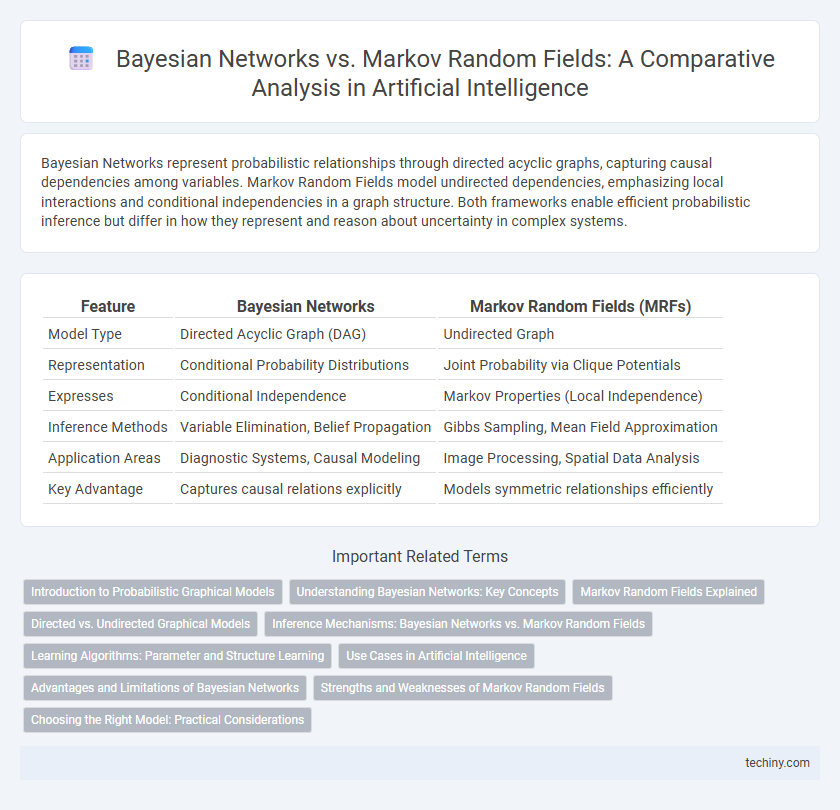
| Feature | Bayesian Networks | Markov Random Fields (MRFs) |
|---|---|---|
| Model Type | Directed Acyclic Graph (DAG) | Undirected Graph |
| Representation | Conditional Probability Distributions | Joint Probability via Clique Potentials |
| Expresses | Conditional Independence | Markov Properties (Local Independence) |
| Inference Methods | Variable Elimination, Belief Propagation | Gibbs Sampling, Mean Field Approximation |
| Application Areas | Diagnostic Systems, Causal Modeling | Image Processing, Spatial Data Analysis |
| Key Advantage | Captures causal relations explicitly | Models symmetric relationships efficiently |
Introduction to Probabilistic Graphical Models
Bayesian Networks are directed acyclic graphs that represent conditional dependencies through directed edges, ideal for modeling causal relationships in probabilistic graphical models. Markov Random Fields use undirected graphs to capture mutual influences among variables without inherent directionality, emphasizing joint probability distributions. Both frameworks provide powerful tools for reasoning under uncertainty by encoding complex stochastic processes in structured, interpretable models.
Understanding Bayesian Networks: Key Concepts
Bayesian Networks represent probabilistic models using directed acyclic graphs where nodes denote variables and edges indicate conditional dependencies, enabling efficient reasoning under uncertainty. Each node is associated with a conditional probability distribution that quantifies the impact of parent nodes, facilitating inference and learning from data. These networks excel in scenarios requiring causal relationship modeling and sequential decision making in artificial intelligence applications.
Markov Random Fields Explained
Markov Random Fields (MRFs) represent a class of undirected probabilistic graphical models used to capture the joint distribution of a set of variables with an emphasis on local dependencies and spatial relationships. Unlike Bayesian Networks, which use directed edges to model causal relationships, MRFs utilize undirected edges to encode symmetric interactions, making them particularly effective for image analysis, spatial data modeling, and computer vision tasks. The Markov property ensures that each node in an MRF is conditionally independent of all other nodes given its neighbors, facilitating efficient modeling of complex, high-dimensional data through cliques and potential functions.
Directed vs. Undirected Graphical Models
Bayesian Networks utilize directed acyclic graphs to represent conditional dependencies between variables, enabling causal inference and efficient probability computations. Markov Random Fields employ undirected graphs to model symmetric relationships and spatial dependencies, making them ideal for vision and spatial data analysis. The choice between directed and undirected graphical models depends on the nature of the problem, with Bayesian Networks emphasizing directionality and causality, while Markov Random Fields focus on undirected, mutual interactions.
Inference Mechanisms: Bayesian Networks vs. Markov Random Fields
Bayesian Networks utilize directed acyclic graphs for efficient inference through variable elimination and belief propagation, enabling causal reasoning and accommodating evidence updates dynamically. Markov Random Fields operate over undirected graphs, employing methods like Gibbs sampling and iterative conditional modes to perform inference in scenarios with symmetrical relationships and spatial dependencies. Both frameworks address probabilistic inference but differ in structure and algorithmic approaches tailored to their respective applications in AI.
Learning Algorithms: Parameter and Structure Learning
Bayesian Networks utilize parameter learning algorithms such as Maximum Likelihood Estimation and Bayesian Estimation to optimize conditional probability tables, whereas structure learning involves score-based methods like BIC and constraint-based algorithms to identify the network's directed acyclic graph. Markov Random Fields employ parameter learning techniques including pseudo-likelihood and contrastive divergence, with structure learning often relying on graphical lasso or neighborhood selection for uncovering undirected graph structures. Both models face computational challenges in structure learning, particularly with high-dimensional data and complex dependencies.
Use Cases in Artificial Intelligence
Bayesian Networks excel in modeling causal relationships and reasoning under uncertainty, making them ideal for diagnostic systems, expert systems, and decision support tools in artificial intelligence. Markov Random Fields are preferred for spatial data analysis and image processing tasks due to their ability to model contextual dependencies without assuming a directional flow. Both frameworks are integral in natural language processing and computer vision, where capturing complex probabilistic interactions enhances prediction accuracy and decision-making.
Advantages and Limitations of Bayesian Networks
Bayesian Networks excel in modeling causal dependencies through directed acyclic graphs, enabling efficient probabilistic inference and intuitive representation of conditional independencies. Their ability to handle missing data and support learning from limited samples makes them advantageous for complex decision-making tasks. However, Bayesian Networks are limited by difficulties in representing cyclic dependencies and may face computational challenges with dense networks or large variable sets.
Strengths and Weaknesses of Markov Random Fields
Markov Random Fields (MRFs) excel in modeling spatial dependencies and undirected relationships, making them highly effective for image analysis and spatial data where context matters. They struggle with capturing causal directions and computationally intensive inference due to the complexity of their undirected graph structures. Despite these challenges, MRFs provide robust frameworks for representing local interactions in complex systems, outperforming Bayesian Networks in scenarios with symmetric dependencies.
Choosing the Right Model: Practical Considerations
Bayesian Networks excel in scenarios requiring causal inference and directional dependencies, making them ideal for expert systems and diagnostic applications. Markov Random Fields perform better in modeling spatial or relational data with undirected dependencies, such as image processing and network analysis. Evaluating data structure, interpretability needs, and computational complexity guides the choice between these probabilistic graphical models for accurate and efficient AI solutions.
Bayesian Networks vs Markov Random Fields Infographic
 techiny.com
techiny.com